Hard skills encompass specialized knowledge in curriculum design, assessment techniques, and classroom technology skills that are essential for instructors to improve student learning.
Popular Instructor Resume Examples
Check out our top instructor resume examples that emphasize key skills like curriculum development, student engagement, and effective communication. These samples will guide you in showcasing your accomplishments to potential employers.
Looking to develop your own impressive resume? Our Resume Builder offers user-friendly templates specifically designed for educators, helping you make a great first impression.
Recommended
Entry-level instructor resume
This entry-level resume for an instructor highlights the job seeker's ability to improve student engagement and satisfaction through innovative curriculum development and effective teaching strategies. New professionals in the education field must showcase their instructional skills, relevant certifications, and commitment to improving learning outcomes, even with limited formal work experience.
Mid-career instructor resume
This resume effectively showcases the applicant's strong qualifications and compelling achievements, signaling readiness for advanced teaching roles. The focus on student engagement, curriculum development, and leadership positions reflects a trajectory of growth and commitment to educational excellence.
Experienced instructor resume
The resume highlights the applicant's success in improving student engagement by 30% and leading workshops with a 25% boost in success rates. Bullet points efficiently organize achievements, benefiting professionals who prefer clear, effective summaries.
Resume Template—Easy to Copy & Paste
Jane Johnson
Brookfield, WI 53011
(555)555-5555
Jane.Johnson@example.com
Professional Summary
Innovative Instructor with 8 years experience in advanced curriculum design, workshop facilitation, and optimizing student engagement, leveraging top educational technology strategies.
Work History
Instructor
EdTech Innovations - Brookfield, WI
January 2020 - December 2025
- Delivered classes boosting pass rate by 30%
- Developed curriculum enhancing engagement by 50%
- Conducted workshops, reducing learning time by 20%
Teaching Consultant
StartSmart Tutoring - Brookfield, WI
June 2016 - December 2019
- Optimized tutorial methods improving scores by 25%
- Aided in retaining 90% of student enrollments
- Crafted training sessions increasing effectiveness
Education Specialist
Learning Tree - Brookfield, WI
June 2012 - May 2016
- Enhanced lesson plans raising engagement by 40%
- Mentored 50+ staff yielding improved delivery
- Orchestrated events boosting visibility by 60%
Languages
- Spanish - Beginner (A1)
- French - Intermediate (B1)
- Mandarin - Beginner (A1)
Skills
- Curriculum Development
- Educational Technology
- Student Engagement
- Workshop Facilitation
- Performance Optimization
- Data Analysis in Education
- Learning Management Systems
- Mentorship
Certifications
- Certified Education Trainer - National Education Institute
- Advanced Technology Facilitator - Tech Educators Association
Education
Master of Education Curriculum Design
University of Washington Seattle, WA
June 2012
Bachelor of Arts Education
Washington State University Pullman, WA
June 2010
How to Write an Instructor Resume Summary
Your resume summary is the first thing employers will see, making it important to create a compelling introduction that sets you apart. As an instructor, you should highlight your teaching experience, subject matter expertise, and ability to engage students effectively. To clarify what makes a strong summary, we’ll look at examples that illustrate successful elements and common pitfalls:
I am an experienced instructor with many years of teaching in various settings. I hope to find a position where I can use my skills and help students succeed. A school that values collaboration and offers training opportunities would be perfect for me. I believe I can make a positive impact if given the chance.
- Lacks specific examples of teaching achievements or areas of expertise, making it vague
- Focuses on personal aspirations rather than highlighting what the job seeker brings to the educational institution
- Uses generic phrases like 'many years' and 'positive impact' without offering concrete evidence or details
Experienced instructor with over 8 years in higher education, focusing on curriculum development and student engagement strategies. Increased student retention rates by 20% through innovative teaching methods and personalized mentoring programs. Proficient in various educational technologies, including Learning Management Systems (LMS) and online assessment tools.
- Begins with a clear statement of experience and area of expertise
- Highlights a specific achievement that showcases the ability to improve student outcomes
- Mentions relevant technical skills that are essential for modern instructional roles
Pro Tip
Showcasing Your Work Experience
The work experience section is essential on your resume as an instructor, containing the bulk of your content. Effective resume templates always prioritize this section to highlight your professional journey.
This part should be organized in reverse-chronological order, listing your previous positions clearly. Use bullet points to detail specific achievements and contributions you made in each teaching role.
To provide clarity, we’ll present a couple of examples that illustrate effective entries for instructors. These examples will showcase what makes a strong work history and what pitfalls to avoid:
Instructor
ABC Learning Center – Los Angeles, CA
- Taught students various subjects.
- Created lesson plans and materials.
- Assisted in classroom activities.
- Graded assignments and provided feedback.
- Lacks specific information about teaching methods or curriculum used
- Bullet points do not highlight any achievements or improvements in student performance
- Focuses on routine duties rather than powerful contributions to the educational environment
Instructor
Learning Academy – Boston, MA
August 2020 - Present
- Develop and deliver engaging lesson plans to over 30 students per semester, fostering a dynamic learning environment.
- Use assessment data to tailor instruction, resulting in a 20% increase in student performance on standardized tests.
- Mentor new instructors, providing guidance on curriculum development and classroom management techniques.
- Starts each bullet with powerful action verbs that highlight the job seeker’s contributions
- Incorporates specific metrics to showcase measurable outcomes of instructional strategies
- Emphasizes relevant skills such as mentorship and adaptability in teaching approaches
While your resume summary and work experience are important, don’t overlook the importance of other sections that contribute to a polished presentation. For more in-depth advice on crafting each part of your resume, be sure to check out our comprehensive guide on how to write a resume.
Top Skills to Include on Your Resume
A skills section is vital for any resume, as it offers immediate insight into a job seeker's abilities and qualifications. This section benefits both job seekers and employers by clearly presenting capabilities that match job requirements.
Combine hard and soft skills to show employers you are the right fit.
Soft skills are essential interpersonal abilities such as patience, adaptability, and encouragement that foster a positive learning environment and improve student engagement in the classroom.
When selecting skills for your resume, it's essential to align them with what employers expect. Many organizations use automated systems to filter out job seekers who lack key resume skills, making it important to showcase the right abilities.
To effectively target your application, carefully review job postings for specific skills that are mentioned. Highlighting these qualifications on your resume will not only attract recruiters but also help you pass through ATS screening processes successfully.
Pro Tip
10 skills that appear on successful instructor resumes
Highlighting key skills on your resume is essential to attract the attention of recruiters looking for qualified instructors. You can find these skills in our resume examples, which give you an edge when applying for teaching positions.
Here are 10 skills you should consider including in your resume if they fit your qualifications and role requirements:
Communication
Adaptability
Classroom management
Curriculum development
Assessment design
Digital literacy
Conflict resolution
Collaborative teamwork
Time management
Creativity
Based on analysis of 5,000+ teaching professional resumes from 2023-2024
Resume Format Examples
Choosing the right resume format is important as it highlights your teaching experience, relevant skills, and educational achievements in a way that resonates with potential employers.
Functional
Focuses on skills rather than previous jobs
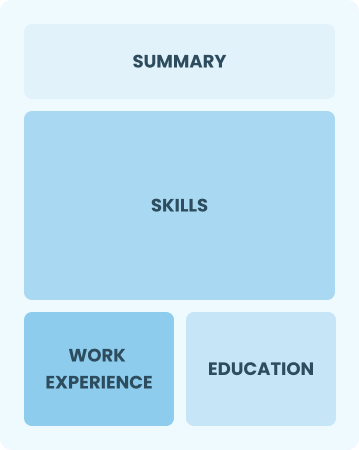
Best for:
Recent graduates and career changers with up to two years of experience
Combination
Balances skills and work history equally
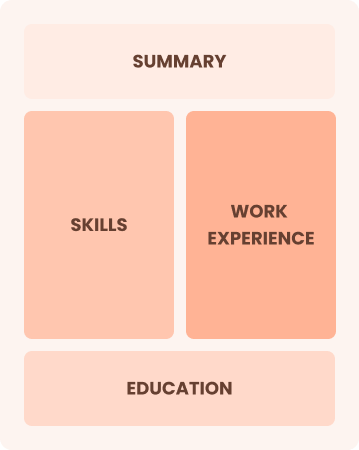
Best for:
Mid-career professionals focused on demonstrating their skills and potential for growth
Chronological
Emphasizes work history in reverse order

Best for:
Seasoned educators leading advanced training in their fields
Instructor Salaries in the Highest-Paid States
Our instructor salary data is based on figures from the U.S. Bureau of Labor Statistics (BLS), the authoritative source for employment trends and wage information nationwide.
Whether you're entering the workforce or considering a move to a new city or state, this data can help you gauge what fair compensation looks like for instructors in your desired area.
Frequently Asked Questions
Should I include a cover letter with my instructor resume?
Absolutely, including a cover letter is essential as it allows you to highlight your unique qualifications and enthusiasm for the position. A well-crafted cover letter can capture a recruiter’s attention and set you apart from other job seekers. For tips on writing one, visit our how to write a cover letter guide or use our Cover Letter Generator to streamline the process effortlessly.
Can I use a resume if I’m applying internationally, or do I need a CV?
When applying for jobs outside the U.S., a CV is often required instead of a resume, especially in Europe and Asia. For guidance on crafting an effective CV, explore our resources on how to write a CV that offer formatting tips and examples to ensure you meet international expectations. You can also review various CV examples to better understand different styles and formats.
What soft skills are important for instructors?
Soft skills like communication, patience, and adaptability are essential for instructors. These interpersonal skills help build positive relationships with students and colleagues, improving the learning environment and supporting effective collaboration in educational settings.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills, such as communication, teamwork, and adaptability from previous roles. These abilities prove your potential to thrive in an instructional role, even if your direct experience is limited. Share concrete examples that link your past successes to key responsibilities in teaching or training settings, showcasing how you can make a positive impact.
Should I include a personal mission statement on my instructor resume?
Yes, including a personal mission statement on your resume is recommended. It effectively conveys your values and career aspirations, making it especially effective for organizations that prioritize cultural fit and share a commitment to their core missions.
How do I add my resume to LinkedIn?
To improve your professional presence as an instructor, add your resume to LinkedIn by integrating key achievements into the "About" and "Experience" sections. This can make it easier for educational institutions and hiring teams to find and connect with you.







