Hard skills are technical competencies such as equipment operation, production scheduling, and basic editing software skill that a production assistant should emphasize.
Popular Production Assistant Resume Examples
Check out our top production assistant resume examples that emphasize essential skills such as organization, communication, and multitasking in fast-paced environments. These samples illustrate how to effectively showcase your contributions to various projects.
Are you ready to build an impressive resume? Our Resume Builder offers user-friendly templates designed specifically for aspiring production assistants to help you shine.
Recommended
Entry-level production assistant resume
This entry-level resume for a production assistant highlights the job seeker's skills in coordinating production workflows and managing resources efficiently, showcasing their achievements in cost reduction and efficiency improvements. New professionals in this field need to demonstrate practical skills and relevant experiences through specific accomplishments, even when they don't have an extensive work history.
Mid-career production assistant resume
This resume effectively showcases qualifications by detailing the job seeker's progressive experience in production roles. The clear articulation of achievements and skills demonstrates readiness for leadership positions, indicating strong potential for managing complex projects and teams.
Experienced production assistant resume
This work experience section illustrates the applicant's robust experience as a production assistant, with notable achievements like increasing production efficiency by 15% and cutting transportation costs by 10%. The bullet-point format improves readability, making it easy for hiring managers to identify key contributions at a glance.
Resume Template—Easy to Copy & Paste
Aiko Wang
San Diego, CA 92114
(555)555-5555
Aiko.Wang@example.com
Professional Summary
Dynamic Production Assistant with 9+ years in multimedia. Skilled in video editing, logistics, and team coordination, ensuring efficient workflows and cost-effective results.
Work History
Production Assistant
Cinemagic Studios - San Diego, CA
January 2022 - December 2025
- Managed equipment inventory, reducing loss by 15%
- Coordinated shoots, ensuring 95% deadline adherence
- Assisted in editing, enhancing video quality by 20%
Film Crew Coordinator
FrameLine Productions - San Francisco, CA
September 2016 - December 2021
- Scheduled 20+ shoots monthly across diverse locations
- Oversaw logistics, reducing transportation costs by 10%
- Trained team, improving efficiency by 25%
Media Production Specialist
Visionary Films - Riverside, CA
August 2013 - August 2016
- Edited 30+ project reels, boosting audience retention
- Set up audio equipment, ensuring 100% error-free sound
- Streamlined workflows, reducing project duration by 20%
Languages
- Spanish - Beginner (A1)
- French - Beginner (A1)
- Italian - Beginner (A1)
Skills
- Video Editing
- Equipment Management
- Project Coordination
- Budget Optimization
- Team Leadership
- Scheduling
- Script Assistance
- Workflow Optimization
Certifications
- Certified Post-Production Specialist - Film Association of America
- Production Safety Coordinator Certified - MPAA Safety Institute
Education
Master of Fine Arts Film Production
University of Southern California Los Angeles, California
June 2012
Bachelor of Arts Media Studies
California State University San Francisco, California
June 2010
How to Write a Production Assistant Resume Summary
Your resume summary is the first impression potential employers will have of you, making it important to craft a compelling introduction. As a production assistant, showcasing your organizational skills and ability to work in fast-paced environments is essential for standing out. The following examples will guide you in understanding effective strategies for writing your own summary and what common pitfalls to avoid:
I am a dedicated production assistant with various experiences in the film industry. I hope to find a job where I can use my skills and help the team achieve its goals. A creative environment that values collaboration and growth is what I’m looking for.
- Lacks specific examples of relevant experience or skills, making it feel generic
- Overuses personal language and focuses on the job seeker’s desires rather than their contributions
- Does not clearly demonstrate how the applicant can add value to potential employers
Proactive production assistant with over 3 years of experience supporting high-profile film and television projects. Contributed to a 20% increase in production efficiency by implementing streamlined communication processes and coordinating logistics for on-set activities. Proficient in Adobe Creative Suite, script breakdowns, and managing equipment inventory to ensure seamless operations.
- Begins with specific number of years of experience and context within the industry
- Highlights a measurable achievement that shows impact on production efficiency
- Mentions relevant technical skills that showcase competencies desired by employers in the production field
Pro Tip
Showcasing Your Work Experience
The work experience section is the focal point of your resume as a production assistant. This area contains the majority of your content, and effective resume templates always prioritize this section.
It's organized in reverse-chronological order, detailing your previous roles. Use bullet points to highlight specific achievements and contributions you've made in each position.
Now, let's look at some examples that illustrate how to craft a compelling work history for production assistants. These examples will clarify what stands out and what should be avoided.
Production Assistant
XYZ Productions – Los Angeles, CA
- Helped on set with equipment.
- Assisted the crew during filming.
- Managed supplies and materials.
- Performed basic office tasks as needed.
- Lacks specific details about contributions to productions
- Bullet points are generic and do not showcase unique skills or accomplishments
- Emphasizes routine duties rather than highlighting any measurable impact on projects
Production Assistant
ABC Studios – Los Angeles, CA
June 2021 - Current
- Assist in the coordination of daily production activities for a variety of television shows, ensuring seamless operations on set.
- Manage equipment and supplies inventory, reducing costs by 15% through efficient tracking and procurement strategies.
- Collaborate with directors and crew to troubleshoot issues promptly, contributing to an on-time project delivery rate of 95%.
- Starts each bullet point with dynamic action verbs that highlight the job seeker's contributions
- Incorporates specific metrics to demonstrate tangible impact on production efficiency
- Showcases essential skills such as coordination and problem-solving directly linked to the role
While your resume summary and work experience are critical components, don’t overlook other sections that also play an important role. Each part of your resume deserves attention to create a strong overall impression. For detailed guidance, check out our comprehensive guide on how to write a resume.
Top Skills to Include on Your Resume
A skills section is important for any effective resume as it quickly communicates your qualifications to potential employers. This area serves as a snapshot of your capabilities, making it easier for hiring managers to see if you're a good fit for the role.
Feature a mix of hard and soft skills to improve your resume.
Soft skills include strong communication, adaptability, and teamwork abilities, which are necessary for collaborating effectively with diverse teams and ensuring smooth production processes.
When selecting your resume skills, it's essential to align them with what employers expect from job seekers. Many organizations use automated systems to filter applicants, so it's important that your resume highlights the relevant skills needed for the role.
To ensure you're showcasing the right abilities, carefully review job postings for insights on which skills are prioritized by recruiters and ATS systems. Tailoring your resume this way will increase your chances of standing out in a competitive job market.
Pro Tip
10 skills that appear on successful production assistant resumes
Improve your resume by incorporating high-demand skills that catch the eye of hiring managers in the production industry. These skills are effectively showcased in our resume examples, providing you with a strong foundation to confidently apply for jobs.
Here are 10 essential skills you might want to feature on your resume if they align with your experience and job specifications:
Attention to detail
Time management
Team collaboration
Problem-solving abilities
Basic technical knowledge
Communication skills
Adaptability
Creativity
Skill in production software
Project coordination
Based on analysis of 5,000+ production professional resumes from 2023-2024
Resume Format Examples
Selecting the correct resume format is important for showcasing your organizational skills, hands-on experience, and growth path as a production assistant.
Functional
Focuses on skills rather than previous jobs
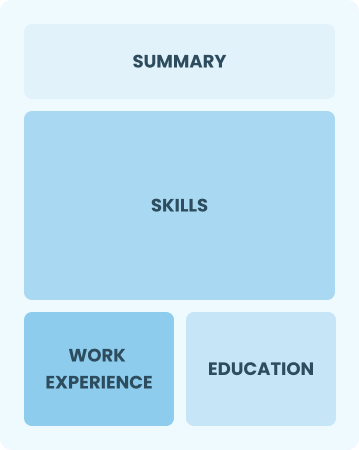
Best for:
Recent graduates and career changers with limited experience in production
Combination
Balances skills and work history equally
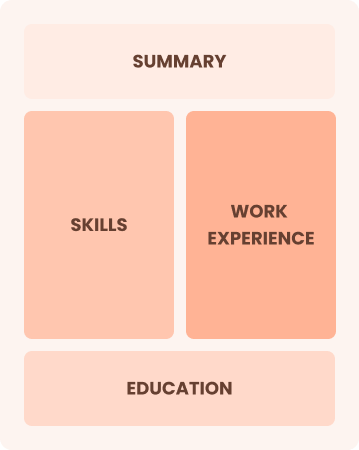
Best for:
Mid-career professionals focused on demonstrating their skills and growth potential
Chronological
Emphasizes work history in reverse order

Best for:
Seasoned production assistants excelling in project management and team leadership
Frequently Asked Questions
Should I include a cover letter with my production assistant resume?
Including a cover letter is beneficial as it highlights your enthusiasm and provides deeper insight into your skills. It allows you to showcase how your experiences align with the position. If you're looking for guidance on how to write a cover letter, our templates can help you craft an effective one quickly and easily. You might also find the Cover Letter Generator useful for creating a personalized letter efficiently.
Can I use a resume if I’m applying internationally, or do I need a CV?
When applying for jobs outside the U.S., use a CV instead of a resume. A CV is often required in many countries and provides a comprehensive overview of your professional history. Explore our resources on how to write a CV for guidance on formatting and creating an effective CV tailored to international standards. You can also look at some CV examples to get a better idea of what works well.
What soft skills are important for production assistants?
Soft skills like communication, teamwork, and problem-solving are essential for production assistants. These interpersonal skills facilitate collaboration with crew members and help navigate challenges on set, ensuring a smooth production process and fostering positive relationships in the fast-paced environment of film and television.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills such as communication, organization, and teamwork when applying for production assistant roles. These abilities illustrate your potential impact on projects despite having limited experience in this field. Provide concrete examples from previous jobs to showcase how your past achievements align with the responsibilities of a production assistant.
How do I write a resume with no experience?
If you're applying for a production assistant role but lack direct experience, focus on showcasing resume with no experience strategies. Highlight relevant skills like organization, teamwork, and familiarity with production tools. Emphasize any internships, volunteer work in media projects, or coursework related to film and television. Your enthusiasm and willingness to learn can leave a lasting impression on employers.
Should I include a personal mission statement on my production assistant resume?
Yes, including a personal mission statement on your resume is recommended. It can effectively showcase your values and career aspirations. This approach is particularly beneficial when applying to organizations that emphasize a strong team culture or have specific guiding principles.







