Hard skills are technical, measurable abilities such as data analysis, statistical methods, and skill in analytical software that are essential for an analyst to effectively interpret data.
Popular Analyst Resume Examples
Check out our top analyst resume examples that demonstrate key skills such as data analysis, problem-solving, and effective communication. These examples can help you highlight your strengths to potential employers.
Ready to build your ideal resume? Our Resume Builder offers user-friendly templates designed specifically for analysts, making the process simple and efficient.
Recommended
Entry-level analyst resume
This entry-level resume for an analyst effectively showcases the applicant's strong analytical skills and accomplishments from previous roles, highlighting their ability to improve data accuracy and efficiency. New professionals in this field should demonstrate technical competencies and problem-solving abilities through quantifiable achievements and relevant project experience, even if their work history is limited.
Mid-career analyst resume
This resume effectively showcases the applicant's qualifications by detailing measurable achievements in data analysis and business intelligence. The structured presentation of skills and accomplishments illustrates a trajectory of growth, signaling readiness for advanced analytical challenges and leadership opportunities.
Experienced analyst resume
This work history section demonstrates the applicant's extensive experience as an analyst, highlighting achievements such as increasing data accuracy by 20% and improving revenue by $500K. The clear bullet-point format allows hiring managers to quickly identify key accomplishments and skills.
Resume Template—Easy to Copy & Paste
Aya Rodriguez
Miami, FL 33105
(555)555-5555
Aya.Rodriguez@example.com
Professional Summary
Dynamic Analyst with 5 years' experience in BI, optimizing data-driven decision-making. Specializes in statistical analysis, increasing process efficiency and leading strategic projects, delivering significant operational improvements.
Work History
Analyst
DataInsights Group - Miami, FL
June 2022 - October 2025
- Increased efficiency by 25% using BI tools.
- Optimized data processes, saving K annually.
- Led data analysis team, resulting in 15% performance boost.
Business Intelligence Specialist
TechSphere Inc. - Jacksonville, FL
February 2020 - May 2022
- Drove 30% increase in report accuracy.
- Implemented dashboard, cutting reporting time 15%.
- Analyzed trends, influencing 1M in investments.
Data Analyst
InsightWorks Solutions - Tampa, FL
October 2018 - January 2020
- Reduced data redundancies by 22%.
- Enhanced data quality, improving user satisfaction 18%.
- Developed KPIs, boosting productivity 10%.
Skills
- Data Analysis
- Business Intelligence
- SQL
- Data Visualization
- Financial Modeling
- Statistical Analysis
- Predictive Analytics
- Project Management
Certifications
- Certified Data Analyst - Data Analyst Association
- Project Management Professional - PMI
Education
Master of Science Data Analytics
Columbia University New York, NY
May 2018
Bachelor of Science Computer Science
University of California Los Angeles, CA
May 2015
Languages
- Spanish - Beginner (A1)
- French - Intermediate (B1)
- German - Beginner (A1)
How to Write a Analyst Resume Summary
Your resume summary is the first impression you make on hiring managers, so it’s important to present your qualifications effectively. As an analyst, showcasing your analytical skills and data-driven insights is essential in this section to grab attention.
Highlight your expertise in interpreting complex data and problem-solving abilities, which are vital traits for success in this field. This helps employers quickly identify your value as a applicant.
To illustrate what makes a strong resume summary, we’ll review some examples that will clarify what works well and what doesn’t:
I am an analyst with some experience in data management. I want to find a job where I can use my skills and grow professionally. A company that values teamwork and offers good benefits would be perfect for me. I believe I can contribute positively if given the chance.
- Lacks specific details about the applicant’s analytical skills and experience, making it too vague
- Emphasizes what the job seeker wants rather than highlighting their unique contributions to potential employers
- Uses generic language that fails to convey any standout qualities or achievements related to analysis
Detail-oriented analyst with 4+ years of experience in data analysis and business intelligence. Improved operational efficiency by 20% through the implementation of a new reporting system that streamlined data collection processes. Proficient in SQL, Excel, and Tableau, with a strong ability to translate complex data into actionable insights for stakeholders.
- Begins with specific years of experience and core competencies relevant to analytics
- Highlights a quantifiable achievement that showcases a significant impact on business operations
- Mentions concrete technical skills that are essential for an analyst role
Pro Tip
Showcasing Your Work Experience
The work experience section is the focal point of your resume as an analyst, where you'll present the bulk of your content. Good resume templates always include this essential section to highlight your professional journey.
This area should be organized in reverse-chronological order, detailing your previous roles. Use bullet points to succinctly describe your key achievements and contributions in each position.
Let's look at some examples that effectively illustrate a strong work history for analysts. These examples will help you understand what works well and what to avoid.
Analyst
Tech Solutions Inc. – New York, NY
- Analyzed data.
- Created reports and presentations.
- Collaborated with team members.
- Assisted with project management tasks.
- Lacks specific accomplishments or outcomes from the analysis work
- Bullet points are overly simplistic and do not highlight unique skills or contributions
- Does not mention any tools or methodologies used in data analysis
Data Analyst
Tech Solutions Inc. – San Francisco, CA
March 2020 - Present
- Analyze large datasets to identify trends and insights, improving marketing strategies that led to a 30% increase in campaign effectiveness.
- Develop interactive dashboards using Tableau, enabling real-time data visualization for stakeholders and reducing reporting time by 50%.
- Collaborate with cross-functional teams to streamline data collection processes, improving data accuracy and reliability across projects.
- Starts each bullet with effective action verbs that clearly convey the analyst's contributions
- Incorporates quantifiable results that demonstrate the tangible impact of the applicant's work
- Highlights collaborative efforts and relevant technical skills essential for success in data analysis
While the resume summary and work experience sections are important, other areas of your resume also demand attention. For more in-depth tips on perfecting each section, explore our complete guide on how to write a resume.
Top Skills to Include on Your Resume
A skills section is an important part of any resume, as it quickly demonstrates your qualifications to potential employers. It helps you stand out by showcasing the technical skills that make you a great fit for the analyst position.
For analysts, highlighting both technical and analytical abilities is key. For example, include expertise in software like Excel, SQL, and data visualization tools such as Tableau to show your strength in interpreting data effectively. However, success truly depends on soft skills like translating complex data narratives for non-technical stakeholders, rigorous attention to detail to ensure accuracy, and the intellectual curiosity to continuously identify new business questions behind the numbers.
Soft skills, including critical thinking, problem-solving, and effective communication, are important for collaborating with teams and translating complex findings into actionable insights.
Choosing the right resume skills is important as it aligns with what employers are seeking in ideal applicants. Many organizations use automated screening systems that filter out applicants lacking necessary skills, making this alignment essential for success.
To ensure you highlight the most relevant abilities, carefully review job postings for insights into desired skills. This approach not only captures recruiters' attention but also improves your chances of passing ATS scans effectively.
Pro Tip
10 skills that appear on successful analyst resumes
Highlighting key skills on your resume not only captures the attention of recruiters but also demonstrates your suitability for analyst roles. Our resume examples showcase these skills in action, allowing you to apply with confidence.
By the way, consider including relevant skills from the following list if they align with your experience and job requirements:
Continuous improvement
Process enhancement
Documentation and reporting
Regulatory compliance
Time management
Process improvements
Decision-making
Ms excel
Report preparation
Analytical thinking
Based on analysis of 5,000+ finance professional resumes from 2023-2024
Resume Format Examples
Selecting the appropriate resume format is important for an analyst, as it allows you to showcase your analytical skills, relevant experience, and career growth in a clear and strong way.
Functional
Focuses on skills rather than previous jobs
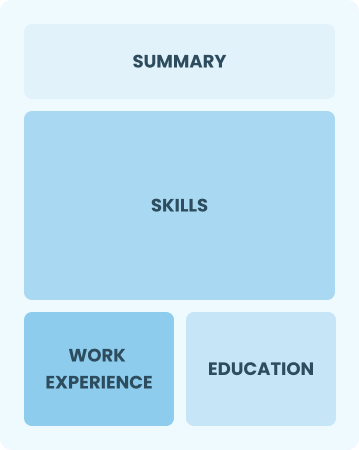
Best for:
Recent graduates and career changers with up to two years of experience
Combination
Balances skills and work history equally
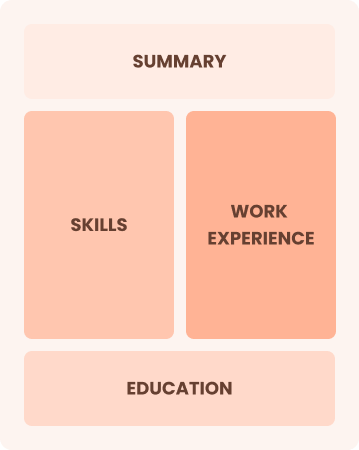
Best for:
Mid-career analysts focused on demonstrating their skills and advancing their careers
Chronological
Emphasizes work history in reverse order

Best for:
Experienced analysts leading strategic initiatives in complex data environments
Analyst Salaries in the Highest-Paid States
Our analyst salary data is based on figures from the U.S. Bureau of Labor Statistics (BLS), the authoritative source for employment trends and wage information nationwide.
Whether you're entering the workforce or considering a move to a new city or state, this data can help you gauge what fair compensation looks like for analysts in your desired area.
Frequently Asked Questions
Should I include a cover letter with my analyst resume?
Absolutely, including a cover letter is a great way to improve your application. It allows you to showcase your personality and clarify how your skills align with the job. For assistance, explore our step-by-step guide on how to write a cover letter or use our Cover Letter Generator for an easy start.
Can I use a resume if I’m applying internationally, or do I need a CV?
When applying for jobs abroad, use a CV instead of a resume, as it's often the standard in many countries. To ensure your CV meets international expectations, explore our comprehensive resources on how to write a CV that can guide you through the process effectively. Additionally, reviewing CV examples can provide valuable insights into proper formatting and content.
What soft skills are important for analysts?
Soft skills like critical thinking, communication, and teamwork are essential for analysts. These interpersonal skills allow you to effectively collaborate with colleagues, present insights clearly, and understand stakeholder needs, ultimately leading to better decision-making and project outcomes.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills such as analytical thinking, communication, and teamwork from past roles. These abilities illustrate your potential to excel in the analyst position, even with limited experience in the field. Use concrete examples to demonstrate how your previous achievements align with job requirements, emphasizing your readiness to add value immediately.
How do I write a resume with no experience?
Aspiring analysts can highlight academic projects, internships, technical skills, and analytical tools skill. Emphasize problem-solving, attention to detail, and critical thinking abilities. When crafting a resume with no experience, your passion for data-driven decision-making can shine through even without extensive job history. Employers value potential and enthusiasm.
How do I add my resume to LinkedIn?
To increase your resume's visibility on LinkedIn, add your resume to LinkedIn by uploading it directly to your profile. You can also highlight essential skills and achievements in the "About" and "Experience" sections. This approach helps recruiters easily find qualified analysts like you, improving your chances of being discovered for relevant opportunities.







