Technical abilities like hard skills in specimen collection, laboratory equipment operation, and data analysis are essential for ensuring accurate test results.
Popular Lab Assistant Resume Examples
Check out our top lab assistant resume examples that emphasize critical skills such as sample preparation, data analysis, and attention to detail. These examples are designed to help you showcase your qualifications effectively.
Ready to build your impressive resume? Our Resume Builder offers user-friendly templates specifically crafted for lab professionals, helping you shine in the job market.
Recommended
Entry-level lab assistant resume
This entry-level resume highlights the job seeker's practical experience in laboratory management and research, showcasing their ability to improve project efficiency and maintain equipment reliability. New professionals in this field must effectively convey their technical skills and collaborative experiences through their resume to attract potential employers, even with limited work history.
Mid-career lab assistant resume
This resume effectively showcases key qualifications by emphasizing hands-on experience and a commitment to safety and efficiency. The clear presentation of skills and accomplishments illustrates the applicant's readiness for advanced lab roles, highlighting their career growth and technical capabilities.
Experienced lab assistant resume
This resume's work history section highlights the applicant's extensive experience in laboratory settings, including conducting over 50 assays monthly with a notable accuracy of 98%. The clear bullet points improve readability, making it easy for hiring managers to quickly identify key achievements and contributions.
Resume Template—Easy to Copy & Paste
John Peterson
Springfield, IL 62705
(555)555-5555
John.Peterson@example.com
Skills
- Sample Management
- Laboratory Techniques
- Equipment Maintenance
- Chemical Analysis
- Protocol Development
- DNA Processing
- Data Documentation
- Safety Compliance
Certifications
- Certified Lab Assistant - National Lab Association
- Advanced Molecular Techniques - BioTraining Institute
Languages
- Spanish - Beginner (A1)
- German - Beginner (A1)
- French - Intermediate (B1)
Professional Summary
Experienced Lab Assistant with skills in sample management, lab techniques, and protocol development. Proven ability to improve accuracy and efficiency in laboratory settings. Fluent in Spanish, with strong teamwork and safety compliance skills.
Work History
Lab Assistant
Precision Labs - Springfield, IL
January 2022 - October 2025
- Managed sample inventory, reduced errors by 25%
- Assisted in over 150 lab tests monthly
- Maintained equipment, cutting downtime by 15%
Laboratory Technician
BioTech Solutions - Springfield, IL
January 2016 - December 2021
- Conducted 200+ chemical analyses annually
- Supervised junior techs, improved efficiency 20%
- Developed protocols increasing accuracy by 10%
Research Assistant
Genomic Research Labs - Chicago, IL
January 2014 - December 2015
- Supported 10+ research projects simultaneously
- Processed 50 DNA samples weekly
- Maintained lab environment, exceeding standards
Education
Master's Degree Biochemistry
University of Washington Seattle, Washington
June 2013
Bachelor's Degree Molecular Biology
Oregon State University Corvallis, Oregon
June 2011
How to Write a Lab Assistant Resume Summary
Your resume summary is the first thing employers see, making it important to create a strong impression that highlights your qualifications. As a lab assistant, you should emphasize your technical skills and your ability to support laboratory operations efficiently.
In this role, showcasing attention to detail, teamwork, and familiarity with lab equipment will set you apart from other applicants. Your summary should reflect those strengths and demonstrate your readiness for the job.
To help you identify effective strategies for writing your own resume summary, consider these examples that illustrate what works well in capturing an employer's interest:
I am a lab assistant with experience in various laboratory settings. I want to find a job where I can use my skills and contribute to the team. A position that offers flexibility and room for advancement is what I am looking for. I believe my background will benefit your organization if given the chance.
- Lacks specific details about relevant skills or experiences related to laboratory work
- Contains excessive personal language rather than focusing on contributions to potential employers
- Emphasizes applicant desires instead of highlighting what they can bring to the role and organization
Detail-oriented lab assistant with 3+ years of experience in clinical laboratory settings, specializing in sample analysis and quality control. Improved turnaround time for test results by 20% through process optimization and efficient workflow management. Proficient in using laboratory information systems (LIS), performing complex assays, and ensuring compliance with safety regulations.
- Begins with specific experience level and area of expertise within the laboratory field
- Highlights a quantifiable achievement that demonstrates impact on operational efficiency
- Includes relevant technical skills that align with the responsibilities of a lab assistant
Pro Tip
Showcasing Your Work Experience
The work experience section is important on your resume as a lab assistant, where you'll have the bulk of your content. Good resume templates always emphasize this important area.
This section should be organized in reverse-chronological order, detailing your past positions. Include bullet points that highlight your specific achievements and contributions in each role to make them stand out.
Now, let’s look at a couple of examples that demonstrate effective ways to present your work history as a lab assistant. These examples will help clarify what works well and what should be avoided:
Lab Assistant
City Medical Labs – San Francisco, CA
- Assisted with lab tests
- Maintained equipment and supplies
- Recorded data in lab reports
- Supported senior technicians in daily tasks
- Lacks specific employment dates
- Bullet points are generic and do not highlight individual contributions
- Emphasizes routine duties rather than effective achievements
Lab Assistant
City Hospital Laboratory – Chicago, IL
February 2020 - Current
- Conduct routine laboratory tests on blood and other specimens, ensuring accuracy and timeliness in results for over 200 patients weekly
- Assist in the development of new testing protocols that improved efficiency by 30%, reducing turnaround times significantly
- Train and supervise junior lab technicians, fostering a collaborative environment that improved team productivity and skill acquisition
- Starts each bullet with action verbs to clearly communicate the applicant's contributions
- Incorporates specific metrics to quantify achievements, lending credibility to the applicant's effectiveness
- Highlights relevant skills such as training and protocol development that align with industry standards
While your resume summary and work experience sections are important, don’t overlook the other important parts that contribute to your overall presentation. For more in-depth advice on crafting a standout resume, explore our comprehensive guide on how to write a resume.
Top Skills to Include on Your Resume
A skills section is an important part of your resume because it lets you showcase your qualifications at a glance. It highlights your strengths and helps potential employers quickly assess if you're the right fit for the lab assistant role.
For this position, focus on both hard skills and soft skills to show employers you're the perfect candidate. While hard skills are crucial for demonstrating your subject matter expertise, soft skills assure employers you have the personal traits to succeed.
Skills such as attention to detail, teamwork, and effective communication are examples of soft skills, which are important for collaborating with healthcare professionals and maintaining high-quality patient care.
When selecting skills for your resume, it's important to align with what employers expect from job seekers. Many organizations use automated screening systems that filter out applicants lacking essential resume skills.
To improve your chances of standing out, carefully review job postings to identify the key skills highlighted by recruiters. Prioritizing the most relevant abilities will help ensure your resume appeals to both hiring managers and ATS systems alike.
Pro Tip
10 skills that appear on successful lab assistant resumes
Highlighting essential skills on your resume can significantly attract recruiters looking for lab assistants. You can see these high-demand skills represented in our resume examples, ensuring you present yourself confidently to potential employers.
Consider adding the following 10 relevant skills to your resume if they align with your background and job expectations:
Attention to detail
Laboratory techniques
Data analysis
Time management
Team collaboration
Quality control procedures
Knowledge of safety protocols
Technical skill with lab equipment
Sample collection and preparation
Effective record-keeping
Based on analysis of 5,000+ administrative professional resumes from 2023-2024
Resume Format Examples
Choosing the appropriate resume format is important for a lab assistant, as it effectively showcases essential skills, relevant experience, and career growth to potential employers.
Functional
Focuses on skills rather than previous jobs
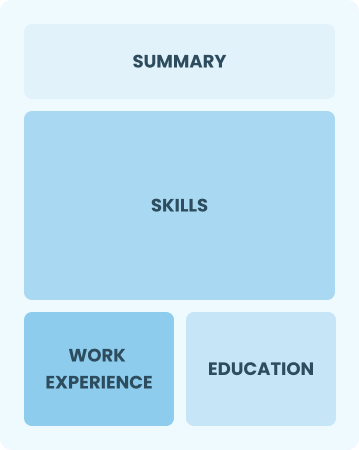
Best for:
Recent graduates and career changers with up to two years of experience
Combination
Balances skills and work history equally
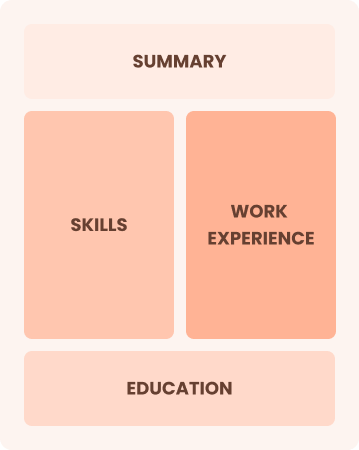
Best for:
Mid-career professionals focused on demonstrating their skills and growth potential
Chronological
Emphasizes work history in reverse order

Best for:
Experienced lab assistants excelling in complex analytical techniques
Frequently Asked Questions
Should I include a cover letter with my lab assistant resume?
Absolutely, including a cover letter can significantly improve your application. It allows you to showcase your personality and detail how your skills align with the job. For tips on crafting an effective cover letter, consider exploring our comprehensive guide on how to write a cover letter or use our Cover Letter Generator for a quick start.
Can I use a resume if I’m applying internationally, or do I need a CV?
When applying for jobs abroad, a CV is often required instead of a resume. This document provides a comprehensive overview of your academic and professional history. To ensure your CV meets the expectations of international employers, explore how to write a CV for guidance on formatting and creation. Additionally, review various CV examples to better understand what international employers might expect.
What soft skills are important for lab assistants?
Soft skills like communication, attention to detail, and teamwork are essential for lab assistants. These interpersonal skills foster effective collaboration with colleagues and ensure accurate data sharing, ultimately improving the quality of laboratory work and patient care.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills such as communication, teamwork, and analytical thinking from previous roles. These abilities prove you can adapt and excel in a lab assistant position, even if your direct experience is limited. Use clear examples that relate your past successes to the tasks you'll handle in this new role.
How should I format a cover letter for a lab assistant job?
To format a cover letter, start by including your name and contact details, followed by the date and the recipient's information. A professional greeting helps establish a connection with the reader. In the body, emphasize your relevant skills and experiences that align with lab assistant roles. Wrap up with a compelling closing statement encouraging further discussion about your application.
Should I include a personal mission statement on my lab assistant resume?
Including a personal mission statement on your resume is recommended as it effectively showcases your values and career aspirations. This approach works especially well for organizations that prioritize alignment with their core mission or value-driven cultures.







