Hard skills for a PhD job seeker include research methodology, data analysis, and academic writing, which are essential for conducting rigorous studies and publishing findings.
Popular PhD Candidate Resume Examples
Discover our top PhD job seeker resume examples that emphasize critical research skills, effective communication, and strong analytical abilities. These examples will help you showcase your academic achievements in a way that captivates potential employers.
Looking to build your own impressive resume? Our Resume Builder offers customizable templates designed specifically for job seekers pursuing advanced degrees, making the process straightforward and efficient.
Recommended
PhD candidate resume
The resume uses a structured format with clear headings and bullet points, improving readability for hiring managers. The choice of resume fonts ensures the job seeker's information is presented concisely while reflecting their attention to detail, creating a strong impression in a competitive job market.
Research fellow resume
This resume effectively integrates critical skills such as quantitative analysis and statistical modeling with relevant work experience. By showcasing these abilities alongside a history of effective research contributions, employers can clearly assess the applicant's expertise and potential for success in advanced research roles.
Doctoral researcher resume
This resume effectively uses bullet points to clearly present the job seeker's extensive experience, making key qualifications easy to spot. The thoughtful spacing between sections and concise descriptions ensure that hiring managers can quickly grasp the applicant's accomplishments and expertise without feeling overwhelmed.
Resume Template—Easy to Copy & Paste
Maria Davis
New York, NY 10003
(555)555-5555
Maria.Davis@example.com
Professional Summary
Dynamic PhD Candidate with 9 years in data analysis and machine learning. Proven record in AI research, efficiency improvements, and project management. Adept in Python, big data, and statistical modeling.
Work History
PhD Candidate
AI Research Institute - New York, NY
January 2023 - December 2025
- Authored 3 journal papers on AI.
- Conducted research on machine learning.
- Supervised 2 undergraduate projects.
Senior Data Analyst
Tech Innovations Inc. - Brooklyn, NY
January 2017 - December 2022
- Improved data processing efficiency by 30%.
- Developed data models for M project.
- Collaborated with 5 departments for analytics.
Junior Data Analyst
SkyNet Solutions - Newark, NJ
January 2014 - December 2016
- Analyzed data for marketing campaigns.
- Generated weekly reports for 2M sales.
- Reduced data entry errors by 15%.
Languages
- Spanish - Beginner (A1)
- French - Intermediate (B1)
- German - Beginner (A1)
Skills
- Machine Learning
- Data Analysis
- Python Programming
- Statistical Modeling
- Big Data Technologies
- Artificial Intelligence
- Research Methodologies
- Project Management
Certifications
- Data Science Professional Certificate - Harvard University
- Certified Machine Learning Specialist - Coursera
Education
Master's Degree Computer Science
Massachusetts Institute of Technology Cambridge, MA
May 2014
Bachelor's Degree Information Technology
University of California, Berkeley Berkeley, CA
May 2012
How to Write a PhD Candidate Resume Summary
Your resume summary is the initial touchpoint for hiring managers, making it essential to craft a compelling introduction that captures attention. As a PhD applicant, you should emphasize your research experience and academic achievements to showcase your expertise in your field. To illustrate effective strategies, the following examples will clarify what makes a powerful summary and what pitfalls to avoid:
I am a dedicated PhD applicant with a strong academic background and research experience. I am seeking an opportunity that allows me to apply my knowledge and contribute positively to the field. A collaborative environment with mentorship opportunities is very important to me. I believe I would be a valuable asset if given the chance.
- Lacks specific details about research accomplishments or methodologies, making it too vague
- Overuses personal pronouns, which may dilute professionalism in formal applications
- Emphasizes what the applicant desires from the role rather than showcasing their unique contributions and skills
Ambitious PhD job seeker with 4 years of research experience in molecular biology, focusing on gene therapy innovations. Successfully published 3 peer-reviewed articles and presented findings at 5 international conferences, improving lab visibility and collaboration. Proficient in CRISPR technology, bioinformatics tools, and statistical analysis software.
- Begins with specific academic level and research focus area
- Highlights quantifiable achievements that showcase contributions to the field
- Mentions relevant technical skills that align with industry expectations
Pro Tip
Showcasing Your Work Experience
The work experience section is important for your resume as a phd job seeker, serving as the primary area where you showcase your academic and research accomplishments. Effective resume templates always feature this section prominently.
This part of the resume should be arranged in reverse-chronological order, detailing your relevant positions and experiences. Use bullet points to highlight key achievements and contributions in each role you've held.
To assist you further, we’ve prepared some examples that illustrate strong entries for the work experience section of a phd job seeker's resume. These examples will clarify what makes an entry effective or ineffective:
PhD Job seeker
University of Science – Cityville, ST
- Conducted research in the lab.
- Attended seminars and workshops.
- Collaborated with peers on projects.
- Wrote papers and presented findings.
- Lacks specific details about research focus or results achieved
- Bullet points do not highlight unique skills or contributions
- Emphasizes routine activities rather than powerful outcomes
PhD Job seeker
University of Science – Cityville, ST
August 2020 - Present
- Conduct independent research in the field of molecular biology, focusing on gene editing techniques that improve crop yield by 30%.
- Collaborate with a team of researchers to publish findings in top-tier journals, contributing to three publications within two years.
- Mentor undergraduate students in laboratory techniques and data analysis, improving their practical skills and academic performance.
- Uses powerful action verbs at the start of each bullet point to highlight achievements
- Incorporates specific metrics such as percentages and publication counts to quantify success
- Demonstrates essential skills relevant to academia and research through detailed descriptions of accomplishments
While your resume summary and work experience are important components, don't overlook the importance of other sections. Each part plays a role in showcasing your qualifications. For detailed guidance on crafting a standout resume, be sure to explore our comprehensive guide on how to write a resume.
Top Skills to Include on Your Resume
A technical skills section is important for succinctly showcasing your qualifications. It helps employers quickly assess if you possess the capabilities they need.
Strengthen your resume with a balanced mix of hard and soft skills.
Soft skills, such as critical thinking, collaboration, and communication, play an important role in effectively presenting research ideas and fostering productive academic discussions.
When selecting skills for your resume, it's important to align them with what employers expect. Many organizations use automated systems to filter out applicants who lack the necessary resume skills for the position.
To ensure your application stands out, carefully review job postings to identify which resume skills are emphasized. This will help you tailor your resume effectively, appealing both to recruiters and ATS systems alike.
Pro Tip
10 skills that appear on successful phd candidate resumes
Highlighting essential skills on your resume is key to attracting the attention of recruiters for PhD applicant roles. You can see these sought-after abilities illustrated in various resume examples, helping you present yourself with confidence.
By the way, consider incorporating relevant skills from the list below that align with your expertise and job expectations:
Research skill
Analytical thinking
Academic writing
Data analysis
Presentation skills
Time management
Collaboration
Problem-solving
Creativity
Project management
Based on analysis of 5,000+ psychology professional resumes from 2023-2024
Resume Format Examples
Selecting the ideal resume format is important for a PhD job seeker, as it showcases your research accomplishments, academic background, and relevant skills in a clear and effective way.
Functional
Focuses on skills rather than previous jobs
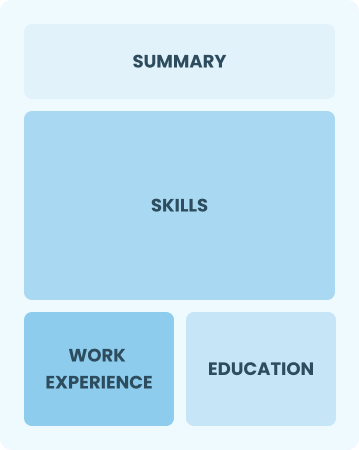
Best for:
Recent graduates and career changers with limited experience in the field
Combination
Balances skills and work history equally
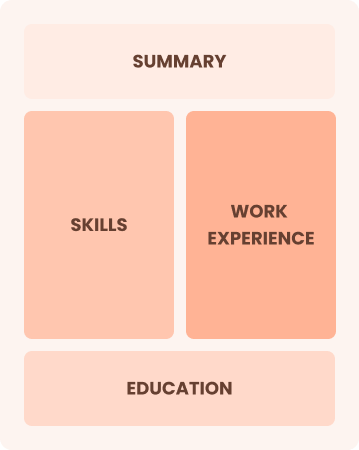
Best for:
Mid-career professionals focused on showcasing skills and pursuing new opportunities
Chronological
Emphasizes work history in reverse order

Best for:
Seasoned leaders excelling in research and innovation
Frequently Asked Questions
Should I include a cover letter with my phd candidate resume?
Absolutely. Including a cover letter is essential as it highlights your unique qualifications and enthusiasm for the position. It gives you an opportunity to elaborate on your resume and make a strong impression. If you need help crafting your cover letter, explore our comprehensive guide on how to write a cover letter or use our Cover Letter Generator for quick assistance.
Can I use a resume if I’m applying internationally, or do I need a CV?
For international job applications, use a CV instead of a resume when applying in countries that expect detailed academic and professional histories. To assist you, we offer resources with CV examples and guidance on how to write a CV that aligns with global expectations.
What soft skills are important for PhD candidates?
Soft skills such as critical thinking, collaboration, and effective communication are essential for PhD. job seekers. These interpersonal skills help build constructive relationships with peers and advisors, fostering a productive research environment and improving the overall academic experience.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills such as research, analytical thinking, and communication. These abilities illustrate your versatility and readiness to succeed in a PhD program, even if your previous experience isn't directly related. Share concrete examples from your past that align with the demands of academic research and collaboration to showcase how you can add value.
How do I write a resume with no experience?
As a PhD job seeker, you can showcase your research projects, academic achievements, and relevant coursework to demonstrate your expertise. Highlight any teaching assistant roles or internships that reflect your skills. To guide you in crafting an effective resume with no experience, focus on your critical thinking, problem-solving abilities, and passion for your field. Your unique background will certainly catch the attention of employers.
How do I add my resume to LinkedIn?
To add your resume to LinkedIn and increase its visibility, upload it directly to your profile or highlight key achievements in the "About" and "Experience" sections. This strategy helps recruiters easily identify qualified phd applicants like you, improving networking opportunities in your field.







