Hard skills include skill in laboratory techniques, data analysis, and equipment operation essential for conducting experiments and interpreting results.
Popular Molecular Biologist Resume Examples
Check out our top molecular biologist resume examples that emphasize critical skills such as experimental design, data analysis, and research collaboration. These examples will help you effectively showcase your accomplishments in the field.
Ready to build a compelling resume? Our Resume Builder offers user-friendly templates specifically designed for professionals in the sciences, making it simple to highlight your expertise.
Recommended
Entry-level molecular biologist resume
This entry-level resume for a molecular biologist effectively highlights the job seeker's research skills and significant contributions in gene analysis and data accuracy. New professionals must demonstrate relevant technical expertise, project accomplishments, and the ability to collaborate effectively in research environments to attract potential employers' attention despite limited experience.
Mid-career molecular biologist resume
This resume effectively presents qualifications, showcasing a strong background in molecular biology and leadership capabilities. The strategic highlighting of research achievements and project management reflects the applicant’s readiness for advanced roles and complex challenges in their field.
Experienced molecular biologist resume
This section showcases the applicant's robust experience as a molecular biologist, demonstrating significant contributions such as improving assay accuracy by 25% and reducing costs by $15K. The use of clear bullet points improves readability, making it easy for hiring managers to identify key achievements quickly.
Resume Template—Easy to Copy & Paste
Olivia Williams
St. Louis, MO 63103
(555)555-5555
Olivia.Williams@example.com
Skills
- Genomic Sequencing
- CRISPR-Cas9
- PCR Optimization
- Biomarker Identification
- Laboratory Management
- Data Analysis
- Scientific Writing
- Team Leadership
Languages
- Spanish - Beginner (A1)
- French - Beginner (A1)
- Mandarin - Intermediate (B1)
Professional Summary
Experienced Molecular Biologist with a focus on genetic research, CRISPR-Cas9, and biomarker identification. Proven track record in laboratory management and scientific communication. Seeking to leverage skills in a research-focused role.
Work History
Molecular Biologist
Genetic Research Institute - St. Louis, MO
January 2022 - October 2025
- Conducted genomic DNA sequencing with 98% accuracy
- Led a team to identify novel biomarkers, increasing efficiency by 20%
- Published 12 peer-reviewed papers on molecular genetics
Research Scientist
BioTech Solutions - St. Louis, MO
January 2018 - December 2021
- Developed CRISPR-Cas9 based gene editing techniques
- Optimized PCR protocols, reducing time by 30%
- Supervised laboratory assistants and interns
Lab Technician
GeneWorks Laboratories - St. Louis, MO
January 2015 - December 2017
- Performed routine DNA extractions, increasing yield by 15%
- Maintained lab equipment, reducing downtime by 25%
- Assisted in the preparation of research manuscripts
Certifications
- Certified Molecular Biologist - American Society for Biochemistry and Molecular Biology
- CRISPR Gene Editing Professional - Genetic Engineering Certification Institute
Education
Master of Science Molecular Biology
Stanford University Stanford, California
June 2015
Bachelor of Science Biology
University of California, Berkeley Berkeley, California
June 2013
How to Write a Molecular Biologist Resume Summary
Your resume summary is the first impression employers have of you, making it essential to craft it carefully. As a molecular biologist, showcasing your technical skills and research experience will set you apart in this competitive field. The following examples will illustrate effective strategies for creating powerful summaries that highlight your qualifications and attract attention:
I am a molecular biologist with extensive experience in research and lab work. I hope to find a position where I can use my skills and contribute positively to the team. A supportive environment that allows for personal growth is important to me. I believe my background makes me a strong applicant.
- Contains vague expressions about experience without detailing specific skills or achievements
- Overuses personal pronouns, making the statement feel self-centered rather than focused on employer needs
- Emphasizes what the applicant desires from a job instead of highlighting how they can benefit the organization
Results-driven molecular biologist with 7+ years of experience in genomics and biotechnology research. Successfully led a project that increased gene sequencing efficiency by 30%, contributing to significant advancements in personalized medicine. Proficient in CRISPR technology, bioinformatics tools, and statistical analysis software, collaborating effectively within interdisciplinary teams to drive innovative solutions.
- Begins with clear years of experience and specific areas of expertise
- Highlights a quantifiable achievement showcasing impact on research efficiency
- Includes relevant technical skills that are important for molecular biology roles
Pro Tip
Showcasing Your Work Experience
The work experience section is important in your resume as a molecular biologist, where you'll present the bulk of your accomplishments. Good resume templates always emphasize this area to showcase your professional journey.
This section should be organized in reverse-chronological order, detailing your previous positions. Use bullet points to highlight significant achievements and contributions you made in each role you held.
Next, we’ll present a couple of examples that illustrate effective work history entries for molecular biologists. These examples will clarify what works well and what pitfalls to avoid:
Molecular Biologist
Genetics Lab Inc. – San Diego, CA
- Conducted experiments in a lab.
- Analyzed data and wrote reports.
- Collaborated with team members on projects.
- Maintained laboratory equipment and supplies.
- Lacks specific employment dates which can affect credibility
- Bullet points do not highlight unique skills or achievements
- Describes routine tasks rather than strong contributions or results
Molecular Biologist
GenTech Laboratories – San Diego, CA
March 2020 - Current
- Conduct advanced genetic research leading to the identification of novel biomarkers for early disease detection, improving accuracy by 30%.
- Develop and optimize PCR protocols which increased laboratory throughput by 40%, significantly reducing project timelines.
- Collaborate with a team of scientists to publish findings in peer-reviewed journals, contributing to the advancement of molecular diagnostics.
- Starts each bullet point with strong action verbs that highlight the job seeker’s contributions
- Incorporates specific metrics demonstrating tangible results from their efforts
- Showcases relevant skills and collaborative efforts that align with industry standards
While your resume summary and work experience are important, don’t overlook other important sections that can improve your overall presentation. For more detailed advice on crafting a standout resume, be sure to check out our complete guide on how to write a resume.
Top Skills to Include on Your Resume
A strong resume must include a skills section to effectively highlight your qualifications. This approach allows potential employers to quickly identify if you possess the essential abilities required for the role.
As a molecular biologist, emphasize both technical skills and analytical capabilities. Highlight tools such as PCR machines, DNA sequencing software, and bioinformatics platforms that demonstrate your skill in modern research techniques.
Soft skills encompass critical thinking, teamwork, and communication, which are important for collaborating with colleagues and conveying complex scientific information effectively.
When selecting skills for your resume, it’s important to align them with what employers expect. Many organizations use automated screening systems that filter out applicants lacking essential resume skills.
To improve your chances of success, carefully review job postings for insights into the specific skills in demand. This approach will help you highlight relevant abilities that resonate with both recruiters and ATS software.
Pro Tip
10 skills that appear on successful molecular biologist resumes
Improve your resume by highlighting essential skills that capture the attention of recruiters for molecular biologist roles. Our resume examples showcase these in-demand skills, empowering you to apply with confidence.
By the way, here are 10 skills you should consider adding to your resume if they align with your experience and job expectations:
Analytical thinking
Attention to detail
Problem-solving
Laboratory techniques
Data analysis
Team collaboration
Knowledge of regulatory standards
Project management
Technical writing
Bioinformatics
Based on analysis of 5,000+ biology professional resumes from 2023-2024
Resume Format Examples
Choosing the right resume format is important for a molecular biologist as it highlights key research skills, relevant experiences, and demonstrates professional growth effectively.
Functional
Focuses on skills rather than previous jobs
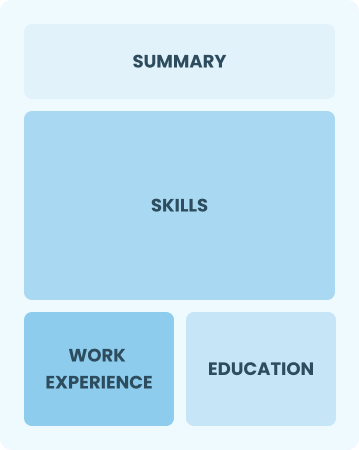
Best for:
Recent graduates and career changers with up to two years of experience
Combination
Balances skills and work history equally
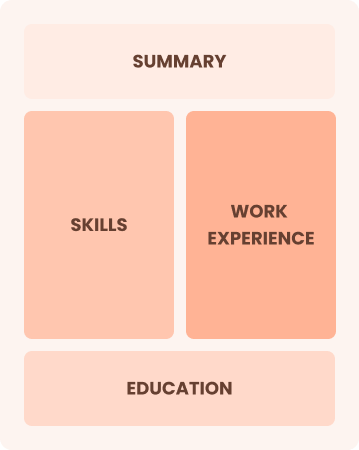
Best for:
Mid-career professionals focused on demonstrating their skills and potential for growth
Chronological
Emphasizes work history in reverse order

Best for:
Experts driving innovative research in molecular genetics and biotechnology
Frequently Asked Questions
Should I include a cover letter with my molecular biologist resume?
Including a cover letter can significantly improve your application by showcasing your personality and qualifications. It allows you to articulate your passion for the position and highlight key experiences. If you need assistance, explore our comprehensive guide on how to write a cover letter or use our Cover Letter Generator to streamline the process.
Can I use a resume if I’m applying internationally, or do I need a CV?
When applying for jobs outside the U.S., use a CV instead of a resume, as many countries favor this format. To assist you, we offer comprehensive resources and CV examples that demonstrate proper formatting and writing techniques to meet international standards. Additionally, if you need guidance on creating your CV, our guide on how to write a CV will be helpful.
What soft skills are important for molecular biologists?
Soft skills, including interpersonal skills like collaboration, communication, and critical thinking, are essential for molecular biologists. These abilities facilitate effective teamwork in research settings and help convey complex scientific concepts clearly, fostering strong relationships with colleagues and improving overall project outcomes.
I’m transitioning from another field. How should I highlight my experience?
Highlight your transferable skills, such as analytical thinking, teamwork, and attention to detail. These attributes are important in molecular biology roles. Even if your background is outside the field, share specific instances where you applied these skills to achieve results. This will showcase your potential and readiness to excel in this new domain.
Where can I find inspiration for writing my cover letter as a molecular biologist?
For aspiring molecular biologists, exploring professional cover letter examples can be incredibly beneficial. These samples provide valuable insights into formatting options, content ideas, and effective ways to showcase your qualifications. Use them as inspiration to craft a compelling narrative that highlights your unique skills and experiences.
Should I include a personal mission statement on my molecular biologist resume?
Yes, incorporating a personal mission statement in your resume is advisable. By doing so, you can effectively highlight your values and career aspirations, which often resonate with organizations that emphasize research-driven cultures and innovation. This method is especially useful when seeking roles at companies dedicated to scientific advancement and collaboration.







